In Fall 2022, Openscapes ran four concurrent Champions Cohorts that included participants from all NOAA Fisheries Science Centers, described in Nationwide Openscapes Training At NOAA Fisheries Science Centers: Facilitating Collaboration, Skill-Sharing, and Open Science. This post is the executive summary shared with Alaska Fisheries Science Center (AFSC) leadership of the work and experiences of AFSC research staff participants.
Update February 21, 2023
- *See also: 2022 Fall Openscapes Champion Cohort Makes Inroads to Open Science, NOAA Fisheries Feature Story
AFSC supported 8 teams from across the center in the 2022 Fall Openscapes Cohort of the Openscapes Champions training program. The teams focussed their energy on a range of important research issues supporting the AFSC mission including fishing tagging, fisheries surveys, marine mammals, genetics, shellfish, fish life history, and survey preparation. The teams consisted of 39 AFSC researchers from 7 AFSC divisions. Three of the 8 teams were cross-divisional. All teams developed their skills for improved analysis workflows, report writing, and manuscript preparation; promoting a culture of inclusivity and collaboration; improving on- and off-boarding processes; and integrated project management. This was achieved in the context of expanding open data science efforts within NOAA and the greater scientific community.
For each team, the training was just the beginning of a longer process. Over the four-week course of the workshop, the teams felt that Openscapes provided them with the support and time to tackle previously intangible tasks, track their progress, and install long-term collaboration and project management practices. While each team tackled very different challenges and goals with unique approaches, they often came to realize that their work needs had universal themes. Teams found new ways to fulfill needs for increased transparency, inclusivity, streamlining workflows, and professional development. The support and enthusiasm of AFSC leadership for AFSC to develop new skills and best practices is critical to AFSC’s continued organizational excellence. Participants in the training also recognized the need for additional training in specific skills and technologies (e.g., R, Python, Jupyter notebooks, Julia, Git and GitHub, Quarto, Posit Connect, reproducible analysis, and collaborative writing). The AFSC can support these needs by 1) identifying and funding training opportunities, and 2) encouraging scientists and supervisors to allocate work time to expanding knowledge and exploring new open data science skills.
As in the first AFSC Openscapes cohort, participants from this cohort found the training transformative. The training created a supportive forum for continued learning and collaboration across programs and divisions. As mentors, we are committed to maintaining the community of support and active learning that was established during the training. We feel confident that AFSC’s continued investment in Openscapes will establish AFSC scientists as leaders in open data science, enhance collaborations across divisions and centers, ensure continuity of long-term research efforts, and improve scientific communication with stakeholders.
The Teams and Their Achievements
The 2022 Fall Openscapes cohort training took place over 5 remote calls from October 5 to November 30, 2022. Cohort calls focused on 1) establishing open science mindset and introducing psychological safety (call 1 digest), 2) publishing and project management on GitHub (call 2 digest), 3) instituting team culture and data strategies (call 3 digest), 4) nurturing open science communities and coding strategies (call 4 digest), and 5) developing each team’s path forward as they continue their open science journeys after Openscapes training (call 5 digest). Participants’ GitHub usernames are provided within each team description to connect them to their Openscapes work.
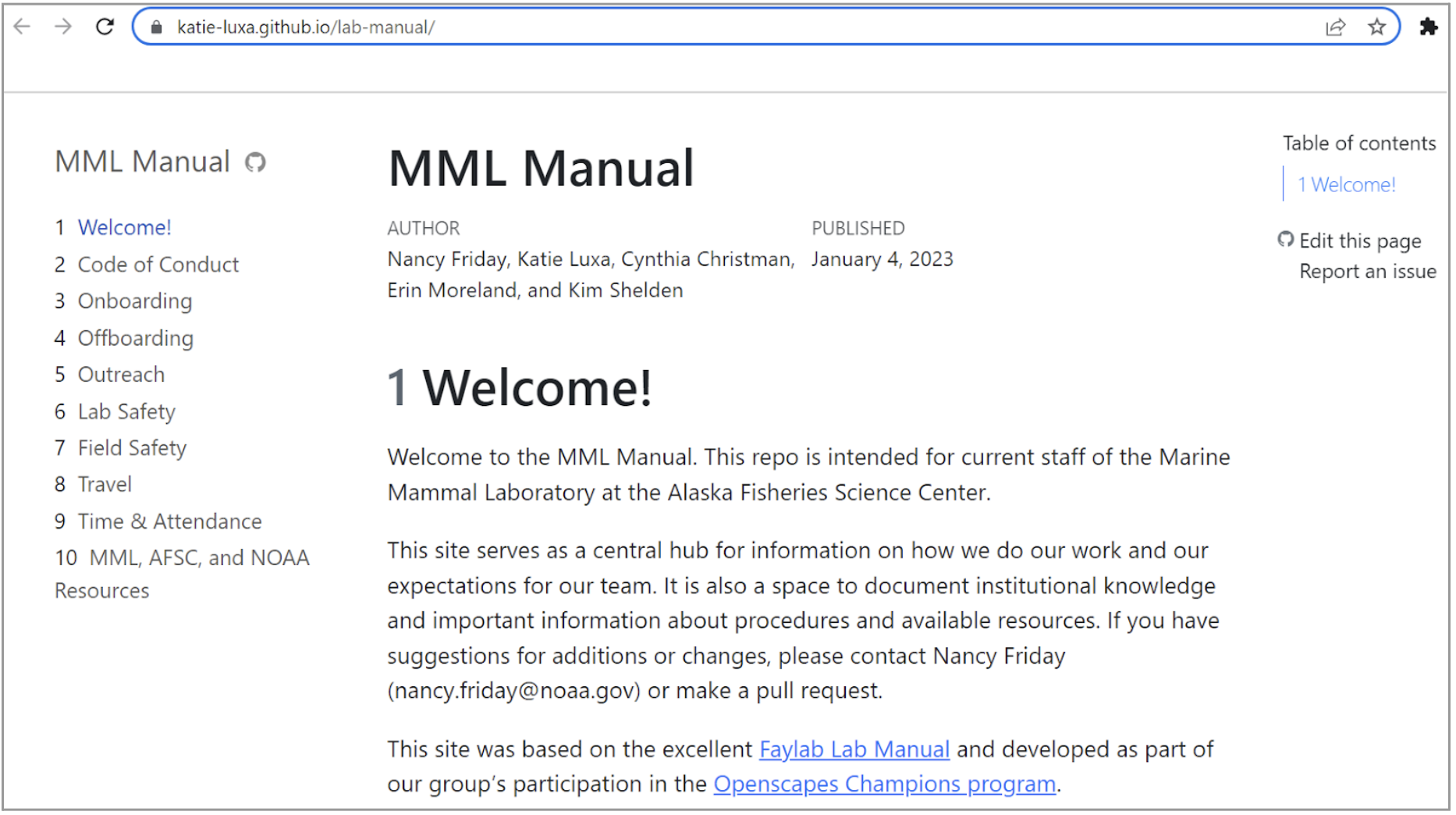
The Marine Mammal Lab/RSST Lab Manual team focused their efforts on developing lab manuals for their teams. This team includes Katie Luxa (@katie-luxa), Erin Moreland (@eem1), Kim Shelden (@kewshel), Nancy Friday (@NAFriday), Cynthia Christman (@ChristmanCL), and Joanna Magner (@joannamagner-noaa; RSST). This cross-division collaboration gave the participants a wider-ranging perspective and an understanding of the universal problems different teams experience. Their lab manual covers on-/off-boarding, outreach, lab safety, field safety, travel, time and attendance, and employee resources. This lab manual was developed by referencing other lab manuals (e.g., Fay lab manual and with the team’s new skills in Quarto, GitHub repo and project management, and GitHub issue tracking. The lab manuals team saw this effort as an opportunity to unbury important lab and employee info from emails in one accessible page. Once complete, this MML lab manual will be moved to the AFSC Marine Mammal Lab GitHub Organization. The members of this team felt that their achievements and end products were more robust and inclusive because of how diversified their team was. The members of this team work across different programs and specialize in research on different species, and could share nuanced perspectives of what different documents and resources should include.
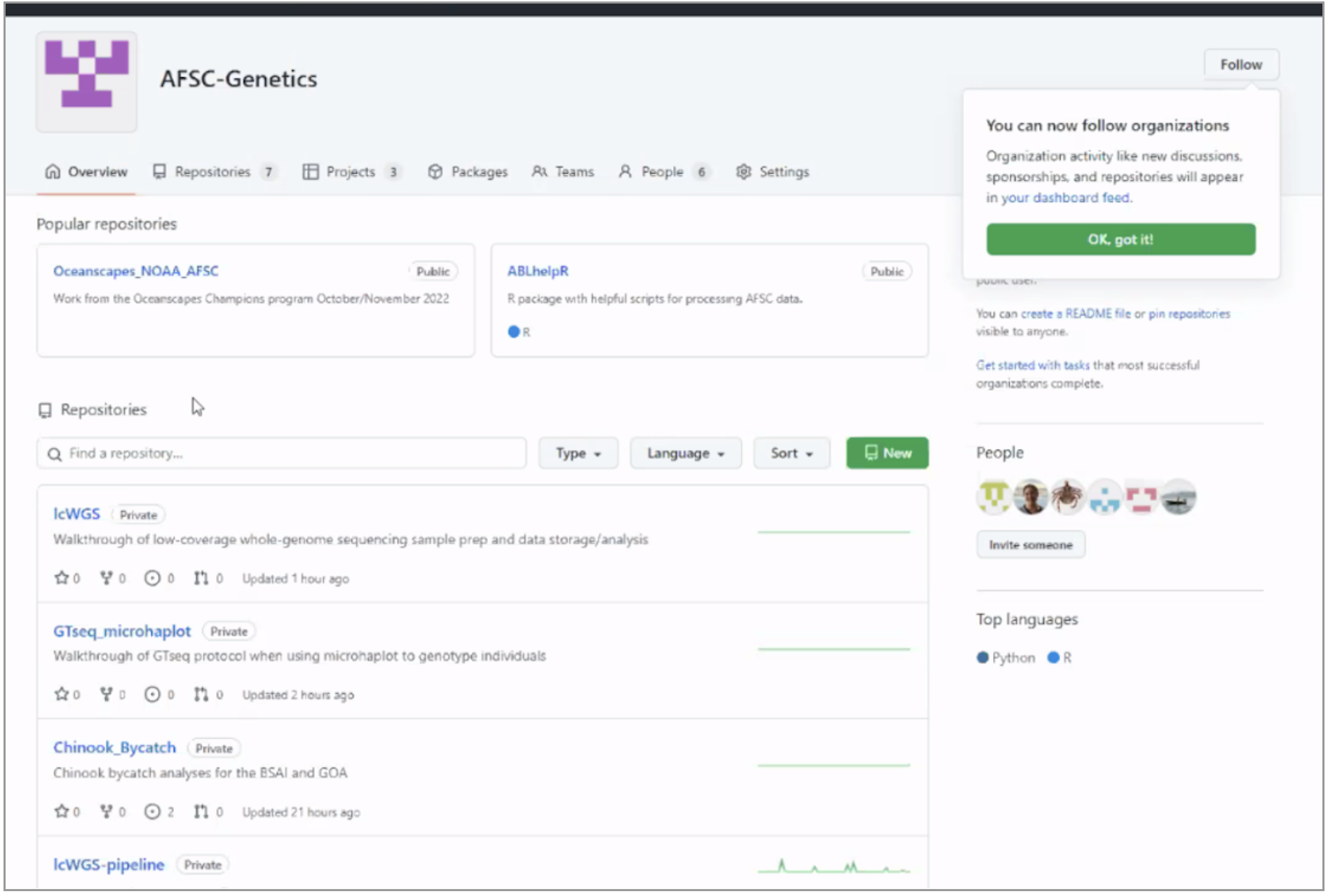
The AFSC Genetics team is motivated to develop a streamlined analysis pipeline. This team is represented by Patrick Barry (@PatrickBarry-NOAA), Diana Baetscher (@DianaBaetscher-NOAA), Sara Schaal (@SaraSchaal), and Claire Tobin (@ClaireTobin-NOAA). The team created a new GitHub organization that allows them to oversee product development, assign issues, and direct overall project management in teams that include postdocs and collaborators. GitHub provides tools for code versioning, documentation, and product testing that were critical for developing this pipeline. Together, they created different GitHub repositories for different projects and initiatives (e.g., sample collection, lab work, data storage) with links to resources and google docs. Members of the team came from a wide range of open science experience levels and worked together to teach and learn from each other through this process.
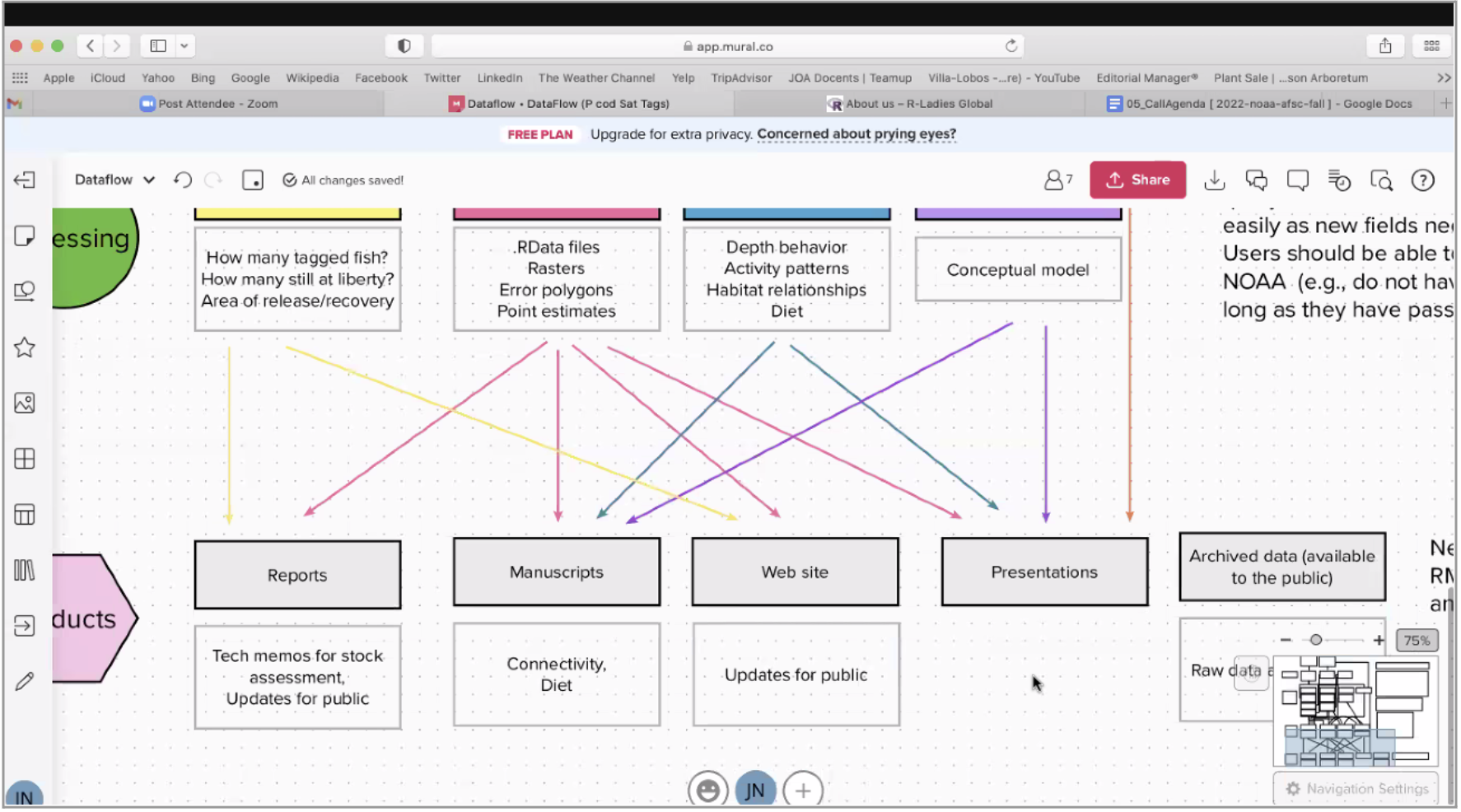
The PCod Tagging and MARVLS team set out on the dynamic task of streamlining multiple data sources to product workflow and diversified data management. This team is comprised of Susanne McDermott (@smcdermo), Julie Nielsen (@jknielsen), Kimberly Rand (@kimberlyrand), Liz Dawson (@liz-dawson-NOAA), Bianca Prohaska (@bianca-prohaska-NOAA), Christina Conrath (@ConrathCl), and Ajith Abraham (@Abraham-7600; OFIS) and ranged across several Pacific cod tagging sub-projects across RACE, the Maturity Assessment, Reproductive Variability, Life Strategy (MARVLS) group, and OFIS. Data from the tagged fish are opportunistically collected when a tag pops off a fish and reaches the sea surface and satellite connection and need to be regularly integrated into reports, websites, and on data sharing platforms. The team used mural boards to assist planning their database workflow and design. This visualization also helped motivate the team, outline what the next steps were, and create and assign tasks. When the team participants came together to work on this project, they were confronted with how similar their project themes and needs were.
The MML Field Report team focused on new methods for working together and developing a ready-for-publication field report. This team includes Kim Goetz (@kimtgoetz), Megan Ferguson (@megancatonferguson), Molly McCormley (@mmccorml), Burlyn Birkmeier (@burlynb), and Amelia Brower (@ameliabrower-noaa). For this project, the team leaned on Eli Holmes’s book chapter code and quarto discussion forums, especially since Quarto is new and quickly evolving. This allowed the team to delegate chapter assignments and other report tasks (e.g., create a new section, edit yaml file, build report) through GitHub issues for team members to work on. The iterative and collaborative nature of the GitHub platform allowed the team to work through the idiosyncrasies of book and page formatting.

The Oyster Monitoring and MML team comprises Peter Mahoney (@pmahoney-noaa), Skyla Walcott (@skylawalcott), Alex Zerbini (@alex-zerbini), Gavin Brady (@GavinMBrady), and Jordan Hollarsmith (@jhollarsmithnoaa). The team is formed of a composite of people from across the center who assisted their peers on their Oyster Monitoring project. For this effort, the team discussed how their universal work needs can be improved by universal lessons and learning GitHub project boards, Quarto, Mural boards (inspired by the PCod tagging team) and RMarkdown. In the Oyster Monitoring GitHub repository, the team worked together to develop the repository and documentation, and they worked with a SeaGrant Fellow to make an interactive quarto report website for the project!
The Groundfish Assessment Program Data Workflow team is working to codify the methods used to calculate survey design-based indices. This is a partnership of the Bering Sea and the Gulf of Alaska/Aleutian Islands groups, including Duane Stevenson (@Duane-Stevenson-NOAA), Lukas DeFilippo (@Lukas-DeFilippo-NOAA), and Sarah Friedman (@SarahFriedman-NOAA), and Zack Oyafuso (@zoyafuso-NOAA). There are many bespoke scripts across the survey groups, for calculations of slightly different flavors of design-based indices, of which running each was often siloed to one person and underwent little to no code review. Expanding on the work already in the GAP Data Products GitHub organization, the team is developing an R package that will consolidate these analyses, interact directly with GAP Oracle databases, and include instructional documentation and vignettes. The package will undergo annual code review and validation, and promote greater collaboration and transparency with data users (e.g., stock assessors). The team relied on GitHub for version control, to post, task, and address issues. In addition, the team led an effort to include the other members of GAP, by initiating working groups to focus on data processes, computations, and future technologies. The team hopes this effort will bring the two survey groups closer together, make these analyses repeatable and transferable, and help develop a future vision for Future Us.
The MACE team was represented by Denise McKelvey (@mckelveyd), Katherine Wilson (@KatherineWilson-NOAA), and David McGowan (@McGowanDW). The program provided the space and guidance for this team to further increase their skills in, and awareness of, the platforms that MACE already uses for collaborative development, maintenance, and sharing of code and standardized reports. The tutorials offered by Eli Holmes (@eeholmes) and the examples of how others have adapted these lessons inspired the team members to envision new ways to collaborate as a team and leverage these tools. The course reinforced the team’s knowledge that Git can facilitate collaborations and increase efficiency by streamlining workflows, tracking issues, and supporting reproducibility. The team appreciated the coding tips that were shared and the cultural change movement that the course inspires: shifting away from work silos and moving towards collaborative and open approaches.
The REFM Spectroscopy Tools for Life History Measurements team comprised Esther Goldstein (@EstherDGoldstein-NOAA), Sandi Neidetcher (@SandiKay), Morgan Arrington (@MorganArrington-NOAA), Irina Benson (@ibens-git), and Beth Matta (@BethMatta-NOAA). This team is working to develop standardized scripts for processing and analyzing spectroscopy data that can be reviewed and shared easily among multiple individuals and centers. This effort is part of a NOAA-wide strategic initiative to develop spectroscopy as a tool for life history measurements (e.g., age, maturity, fish condition). The team realized the need to shift away from siloed scripts used by individuals for specific projects to a generalized open-source format. The team discussed how their work can be improved by using GitHub repositories to review and share scripts, GitHub projects to track issues to assign tasks, and RMarkdown/ Quarto for generating reports more efficiently. The team worked together to develop a GitHub organization, their first repository, practice reviewing scripts, and address issues as a team.
Conclusion
The NMFS Openscapes effort has received some notoriety. Josh London and others presented the cohorts’ successes with Openscapes at the Earth Science Information Partners Conference (ESIP) on January 26, 2023. There is interest across AFSC in further Openscapes training and this may be enabled by a 3-year grant proposal (co-PI’s Julie Lowndes and Eli Holmes) that would further advance this movement toward more open science in NOAA Fisheries. We’re excited to see how things develop in 2023 alongside the White House’s Year of Open Science initiative.