This is a guest blog by Marcus Beck that describes how the Tampa Bay Estuary Program (TBEP) is embracing open science principles and tools to create better science in less time. Marcus, along with TBEP Executive Director Ed Sherwood, both attended the NCEAS Open Science for Synthesis workshop in July 2017 at Santa Barbara. Marcus, Ed, and the rest of the TBEP team are working to bring open science to improve the management of Tampa Bay and extend their applications to the broader network of the 28 National Estuary Programs. This blog describes one application of automating a routine reporting document that builds on the data and tools developed throughout the 30 year history of TBEP.
Update July 13 2021: Check out TBEP’s Data Management SOP( Standard Operating Procedures)!
Those of us working in coastal environments are likely familiar with the success story of Tampa Bay. Like many surface waters in the US prior to the Clean Water Act, Tampa Bay was impacted by nutrient pollution from untreated wastewater and surface runoff. Through a coordinated regional effort of environmental professionals, utility operators, and local politicians, nutrient loads to the Bay have been reduced by ~2/3 from 1970s levels and seagrasses have recovered to a 1950s benchmark extent.
The Tampa Bay Estuary Program (TBEP) has been a key facilitator among the many local partners that have an interest in the region’s natural resources. TBEP is one of 28 estuaries in the EPA’s National Estuary Program that focuses on “estuaries of national significance” to provide place-based solutions to managing these complex systems. This includes establishing benchmarks for restoration goals and tracking the status and trends of key indicators of environmental health.
TBEP uses a water quality report card that summarizes annual progress in achieving core programmatic goals for the Tampa Bay estuary. This product communicates to partners what needs to be done to ensure water quality targets are met to support seagrass growth. However, a new report is needed each year that requires considerable effort to manually compile the data, synthesize the results, and produce the final document.

This blog describes an open science approach developed by TBEP to automate creation of the annual water quality report. The methods leverage many open source tools that are freely available and easily modified for customizable report generation. A broad overview of the approach is provided to encourage others to consider how these open science workflows can lead to better, kinder science in less time.
Why do we need an open science workflow?
Report cards are important communication tools for many resource management agencies. For years, the TBEP report was compiled by hand, by one staff member using external data from our partners. This process has issues, for instance:
- Wasted time each year creating the report card by hand;
- Increased risk of introducing errors into the results through manual analysis steps and workflows; and,
- Closed methods limiting reproducibility, sharing and collaboration with others.
These issues can lead to increased costs, lack of trust by decision-makers, and inhibition of collaboration, all of which are bothersome for publicly-funded agencies. This closed workflow is shown below:
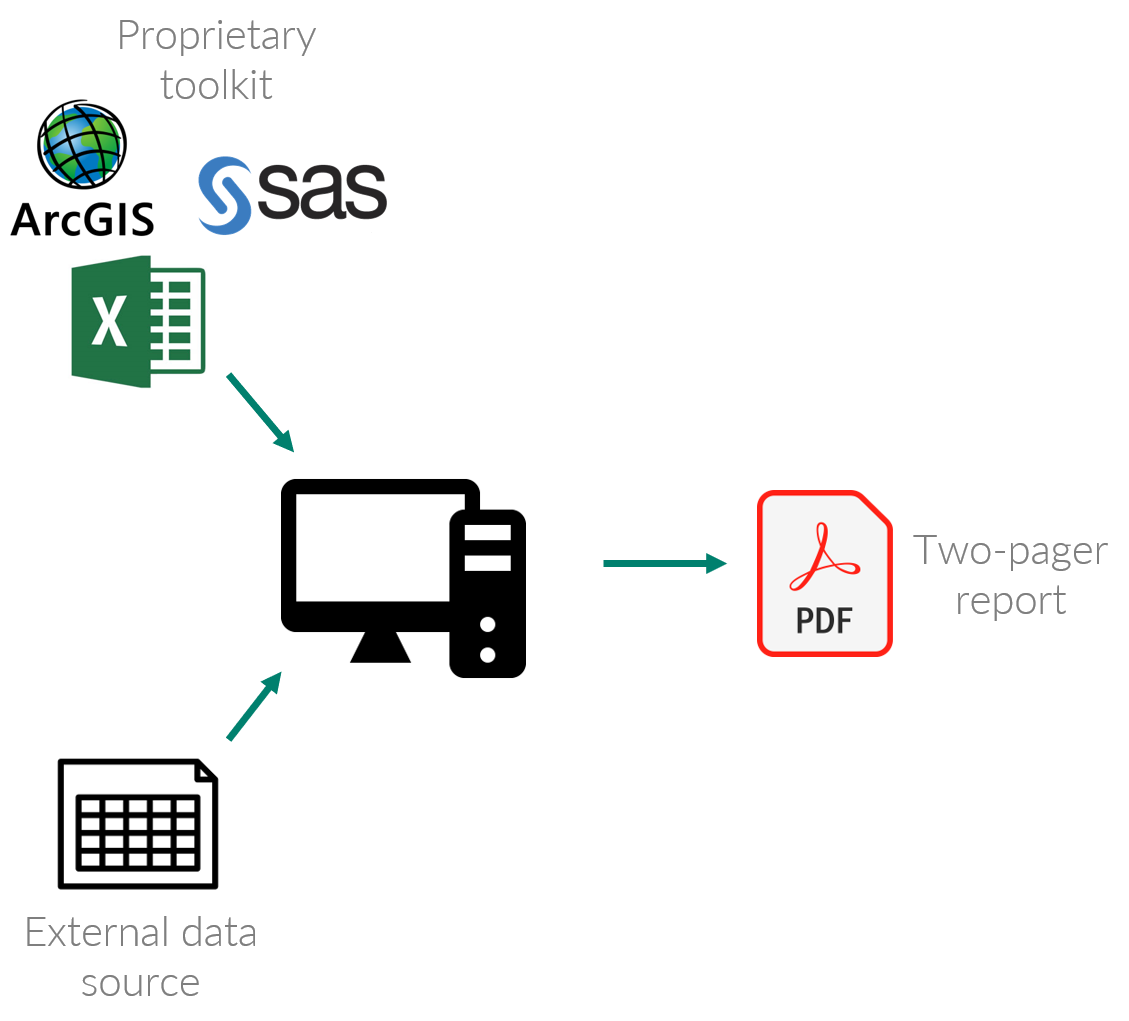
Although the final pdf is a great tool to communicate management needs (and the proprietary tools have their benefits), the report generation process needed to be improved.
Developing an open science workflow
To address inefficiencies of a closed workflow, TBEP has been adopting open science principles to create reporting products that are transparent, efficient, and reproducible. The figure below shows an expanded workflow using open science tools.
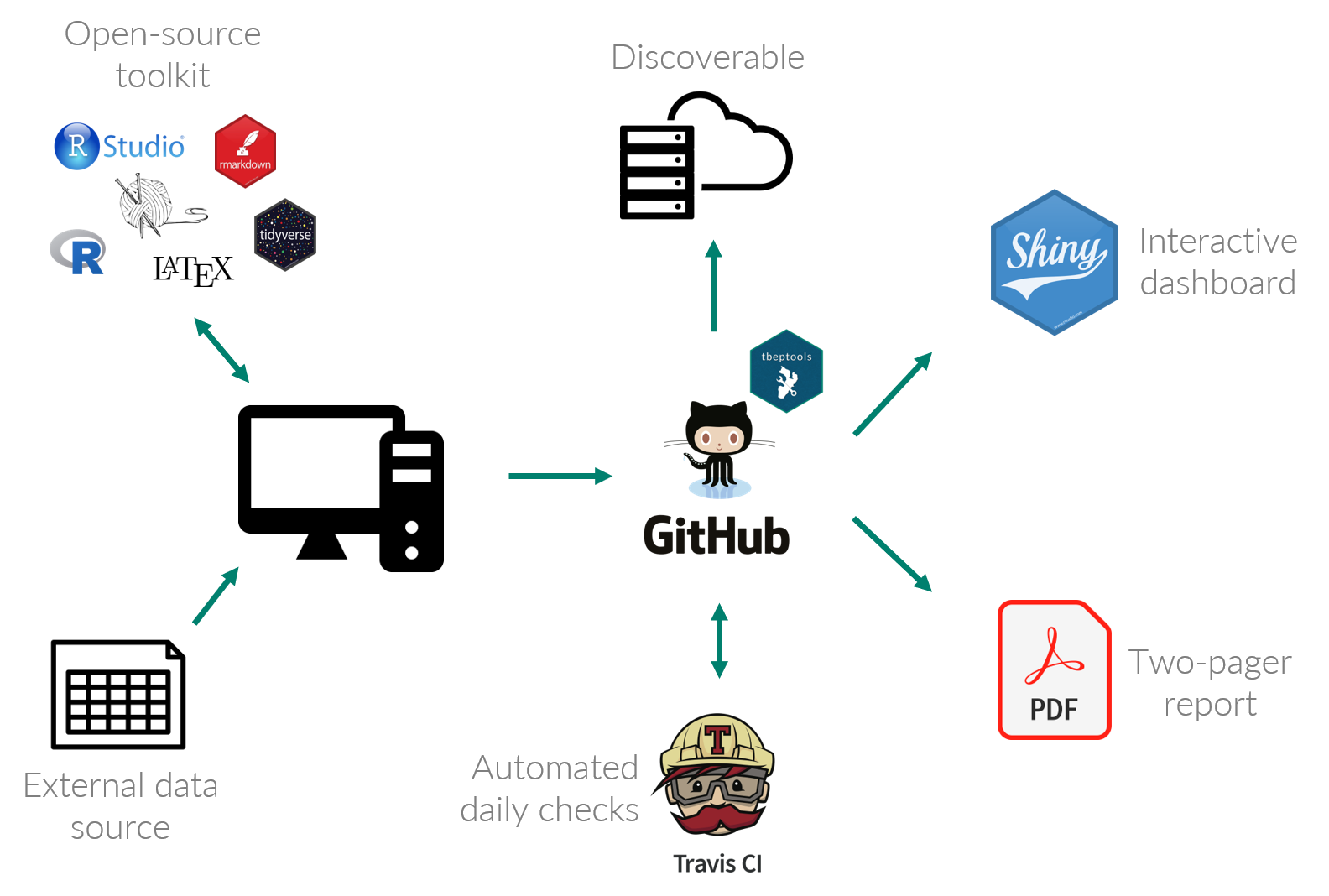
There are several additional tools that make this workflow an improvement over the closed version, such as:
- Use of an open-source toolkit that taps into the broader RStats community;
- Code hosting, sharing, and community development on GitHub;
- Data synthesis and plot generation using the tbeptools R package;
- Continuous integration (CI) using Travis to automate daily checks; and,
- Expansion of reporting tools and science communication, including R Shiny dashboards.
This new open science workflow is similar to the closed version from end to end (i.e., data from partners are used, reports are generated), but several key steps improve how the report is generated to increase efficiencies and expose the underlying methods for reproducibility. Using open-source tools from the RStats community allows us to benefit from the collective knowledge of this dynamic and incredibly welcoming group. Hosting code on GitHub makes the methods discoverable and accessible by others, including our tbeptools R package that includes all methods to read, analyze, and plot data from our external partners. Integration with Travis provides a safety check that our code is running as intended AND allows for daily builds to make sure the most current report is up to date with new data published by our partners. Finally, we use Shiny to create interactive dashboards that allow our partners to engage with the data in a more immersive environment for decision-making and science communication.
Automating the process
Developing the open workflow got us most of the way to achieving open science bliss. An important part of this workflow was including automated builds using the Travis CI service. In the RStats community, Travis is commonly used to check R package builds as a safety net to make sure CRAN guidelines are followed and the package can be installed without any issues. This continuous service is automatic and saves the developer from having to run these checks each time the package is updated. You’ve probably seen those nifty badges on GitHub that indicate if Travis builds were successful or not.
However, Travis can be used for much, much more, including custom builds for websites and highly specific testing services depending on the needs of the developer. We developed our own custom workflow to make use of these services to automatically build the water quality report card. The external data are regularly updated online by our friends at Hillsborough County EPC as part of the Bay’s long-term ambient water quality monitoring program. We wanted to use Travis to build the report card as data are updated, without having to manually create it each time.
How it’s done
All of the pieces that we use to automate creation of the report card live on a GitHub repository here. A simple build script is run daily by Travis to create the final PDF. Within the build script, two .Rnw documents include LaTeX and R code to generate the front and back of the report card. The R code uses functions from the tbeptools R package to import the most current data, summarize the observations, and generate the plots used in the report card. These .Rnw files are then converted to PDF using the tinytex package.
Each day, Travis runs the build script to generate a new PDF. If the build is successful (green badge), it pushes the changes to GitHub so that the new PDF can be hosted on the TBEP website. Any time the build is not successful (red badge), we get an email from Travis telling us something is wrong that we need to fix. This way, we only need to pay attention unless something breaks, allowing us to spend time on other things.
An expanded version of the open science workflow from above shows how the Travis process works:
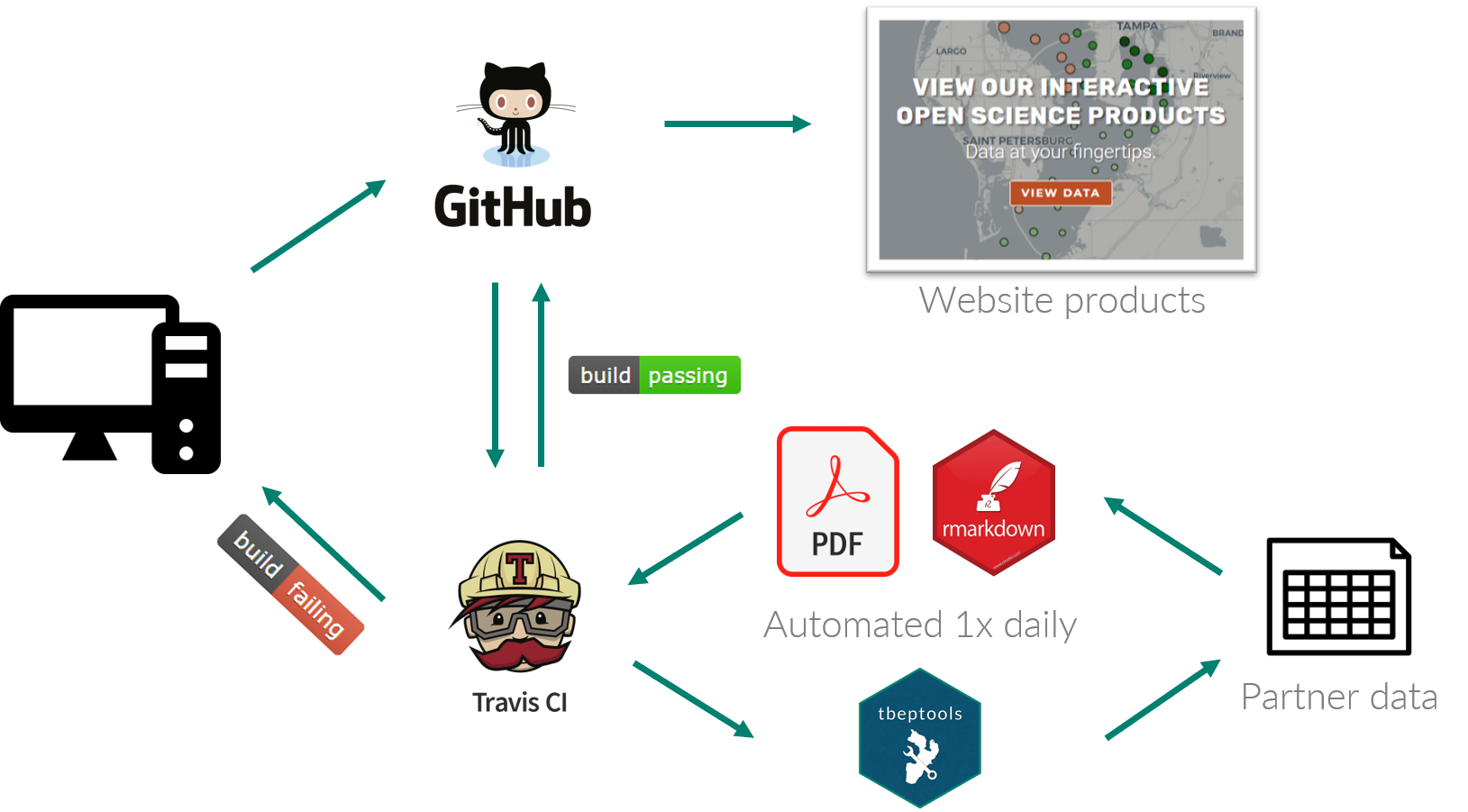
The Travis YAML file in our repository contains the necessary instructions of what to do each day to create the report. There’s a lot to say about modifying the YAML file, but briefly, we use it to specify the package dependencies, how the build is executed, and how it communicates with GitHub to push the changes back when the build is successful.
It’s also worth mentioning that we chose to use Sweave .Rnw files instead of RMarkdown because we needed absolute control over the style and placement of content in the PDF. RMarkdown is great as a simple-to-use tool for generating dynamic documents, but Markdown syntax is purposefully designed for simplicity and does not allow the level of precision we needed for the elements in our report card. This was especially important as it relates to the style of the “old” report card that our partners are used to receiving.
Final thoughts
This workflow was developed as a proof of concept to see if it was reasonable for automating our routine reporting products. We’re in the process of testing and refining this workflow on other products (references library, field data plotting). Although there is substantial overhead in creating the process, we now have a quicker and reproducible approach to delivering decision-support tools that affect the health of Tampa Bay. We are all time-limited, and this workflow reduces the tedium of manually creating these reports each time they are needed.
This workflow is also dependent on the strong technical foundation for evaluating water quality in Tampa Bay and the invaluable work by our partners in collecting/curating the underlying data. This work would not have been possible without the past research, partner initiatives, and community of collaboration that have been the foundation of TBEP and the region’s success in improving the estuary. Having a direct online connection to the raw data is also a necessary component - and on this front, there’s much more work to be done to ensure this information and all of our partner data are Findable, Accessible, Interoperable, and Reusable (FAIR).
Finally, these tools and concepts are generalizable to other locations with similar needs. Open science methods emphasize reproducibility and technology transfer as important benefits that come with investments in developing these tools. The 28 estuary program collective in the US are beginning to embrace these tools so that they can efficiently report on routine estuarine indicators on a broad national scale. This will facilitate shared learning and experiences by using a common and standardized set of reporting products created with open science principles at the core.
Learn more:
- R introduction to using Travis: link
- Setting up Travis CRON jobs: link
- Getting Travis to talk to GitHub: link
- tbeptools R package: link
- TBEP GitHub group web page: link
- TBEP data visualization page: link
- #TampaBayOpenSci